|
Tools.
During the development of this service, the following tools, which have not been incorporated into the final version of the website, were developed.
They are provided due to their value as independent tools and as code sources:
- Oligonucleotide frequencies calculator.
Although in the final version of the website the types of oligonucleotides computed are limited to tetra- to octanucleotides,
we did check additional types of frequencies during its development. This tool allows the retrieval of all of them for one or for both strands of DNA.
The program contains the basic code used by the main program to compute oligonucleotide frequencies and will be fully functional when placed
in a PHP-enabled server.
- Compare oligonucleotide frequencies among sequences.
This tool will compare oligonucleotide frequencies of input sequences, obtain the distances, and generate a dendrogram.
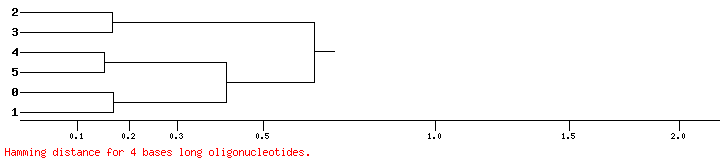
Example of dendrogram generated by this tool.
- Chaos Game Representation (CGR) and CGR of oligonucleotide frequencies (FCGR) .
Comparing oligonucleotide frequencies from different sequences and computing distances among them may be a difficult procedure to undertand.
CGR images are two-dimensional images generated from the primary genome sequences so that they represent the genomes.
They are composed of a huge number of dots, and it is easy to recognize that the density of the dots is variable within the image.
As a consequence, square areas of different sizes can be distinguished in the images. The density of dots in the squares corresponds
to the occurrences of particular oligonucleotides in the genomic sequence (smaller square areas correspond to longer oligonucleotides).
The comparison performed by this website may be understood as a comparison of CGR images. In the case that the CGR images are similar,
the sequences (and the organisms) are similar, while very different CGR images mean the genomes are different.
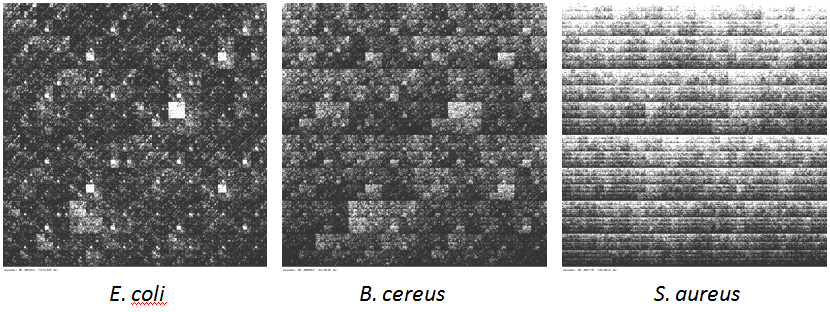
Sample Chaos Game Representation images generated by this tool for three bacteria.
When CGR images are generated for different strains within the same species, images are extremely similar to each other.
|
